The peptidoglycan cell wall supplies a vital protecting barrier in nearly all micro organism, defining mobile morphology and conferring resistance to osmotic stress and different environmental hazards.
The precursor to peptidoglycan, lipid II, is assembled on the internal leaflet of the plasma membrane. However, peptidoglycan polymerization happens on the outer face of the plasma membrane, and lipid II should be flipped throughout the membrane by the MurJ protein earlier than its use in peptidoglycan synthesis. Due to its central function in cell wall meeting, MurJ is of elementary significance in microbial cell biology and is a first-rate goal for novel antibiotic improvement.
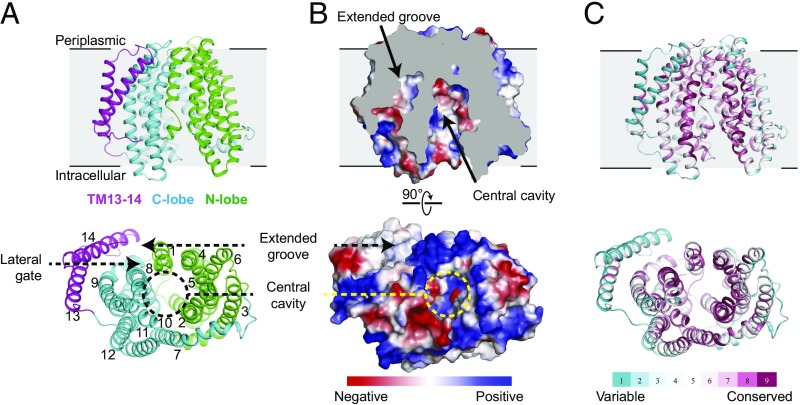
However, comparatively little is thought relating to the mechanisms of MurJ operate, and structural knowledge for MurJ can be found solely from the extremophile Thermosipho africanus Here, we report the crystal construction of substrate-free MurJ from the gram-negative mannequin organism Escherichia coli, revealing an inward-open conformation.
Taking benefit of the genetic tractability of E. coli, we carried out high-throughput mutagenesis and next-generation sequencing to evaluate mutational tolerance at each amino acid in the protein, offering an in depth practical and structural map for the enzyme and figuring out websites for inhibitor improvement.
Lastly, via the use of sequence coevolution analysis, we determine functionally vital interactions in the outward-open state of the protein, supporting a rocker-switch mannequin for lipid II transport.
Dictyostelium as a Model to Assess Site-Specific ADP-Ribosylation Events.
The amoeba Dictyostelium discoideum is a single-cell organism that may endure a easy developmental program, making it a superb mannequin to review the molecular mechanisms of cell motility, sign transduction, and cell-type differentiation.
A range of human genes which are absent or present restricted conservation in different invertebrate fashions have been recognized on this organism. This consists of ADP-ribosyltransferases, often known as poly-ADP-ribose polymerases (PARPs), a household of proteins that catalyze the addition of single or poly-ADP-ribose moieties onto goal proteins.
The genetic tractability of Dictyostelium and its comparatively easy genome construction makes it doable to disrupt PARP gene mixtures, along with particular ADP-ribosylation websites at endogenous loci.
Together, this makes Dictyostelium a lovely mannequin to evaluate how ADP-ribosylation regulates a range of mobile processes together with DNA restore, transcription, and cell-type specification. Here we describe a spread of methods to review ADP-ribosylation in Dictyostelium, together with analysis of ADP-ribosylation occasions in vitro and in vivo, along with approaches to evaluate the practical roles of this modification in vivo.
